ScaleBio Single Cell Methylation Kit
SINGLE CELL DNA Methylation – New and improved!
Advancing DNA methylation
to single cell resolution
Scale Bio’s Single Cell Methylation Kit revolutionizes epigenetics research as the first commercial solution for detecting single cell DNA methylation states. Transform your understanding of biology by exploring epigenetic complexity at a cellular level.
Request a Quote Download Product Sheet
NEW and improved version v1.1
We are excited to announce the release of the Single Cell Methylation Kit v1.1, introducing a more streamlined and flexible workflow to make your single cell epigenetics studies even easier.
Workflow steps have been optimized to reduce nuclei input requirements by 60% and enable target enrichment protocols. Additionally, the kit configuration has been updated to reduce pipetting steps and increase pipetting volumes, with the added benefit of longer fixed nuclei storage times and the introduction of additional safe stopping points.
View the Biofx working group sessions
High-resolution Epigenetic Profiling
Illuminate cell type-specific methylation signatures
·Powerful Chemistry
Interrogate hundreds of thousands of CpG and CH sites per cell
·Scalable Insights
Capture thousands of cells per run with seamless sample multiplexing
Target Enrichment Compatibility
Maximize your sequencing budget by focusing on specific regions of interest

Dr. Federico Gaiti
Princess Margaret Cancer Centre
The Single Cell Methylation Kit from Scale Biosciences has been transformative for our research, particularly in studying the epigenomics landscape of normal and malignant cells in human samples. This innovative product allows researchers to profile a significantly larger number of cells (tens of thousands) per sample compared to other single-cell methylation assays, delivering highly reproducible, high-quality data with extensive cytosine coverage at the single cell level.
Key advantages
True high-resolution epigenetics
Methylation is a heritable and stable epigenetic mark making it ideal for robust detection of epigenetic complexity in tissues. Detect single cell methylomes from thousands of cells with ease with our kitted solution, without hassle of complex, custom protocols or unvalidated reagents.
Discover cell type-specific methylation patterns
Most methylation data today is from bulk-level analysis. Break through the noise and unmask methylation patterns on a cell type-specific level.
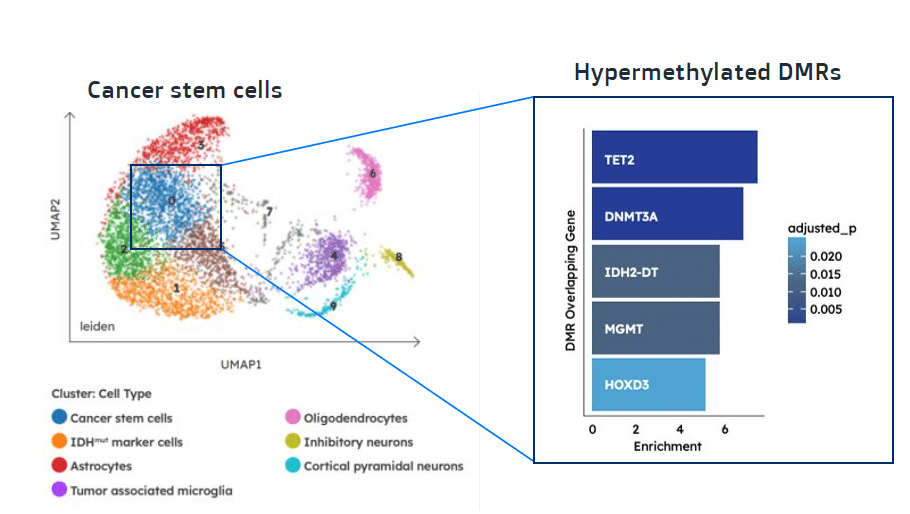
Single cell analysis enables detection of differentially methylated regions specifically in cancer stem cells. Download App Note
Focus on what matters most
Maximize your sequencing budget and expand the number of samples profiled by generating target-enriched libraries, without worrying about leaving important CpGs behind.
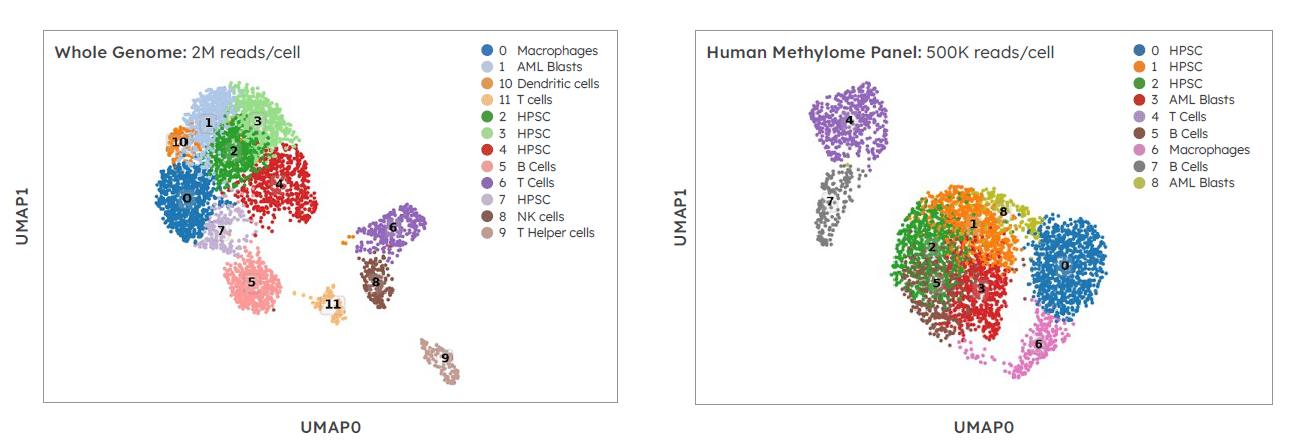
Target enrichment captures similar PMBC cell types as whole genome single cell methylation data with 75% fewer sequencing reads. Targeted single cell methylation library generated using Twist Human Methylome Target Enrichment panel.
Streamlined workflow
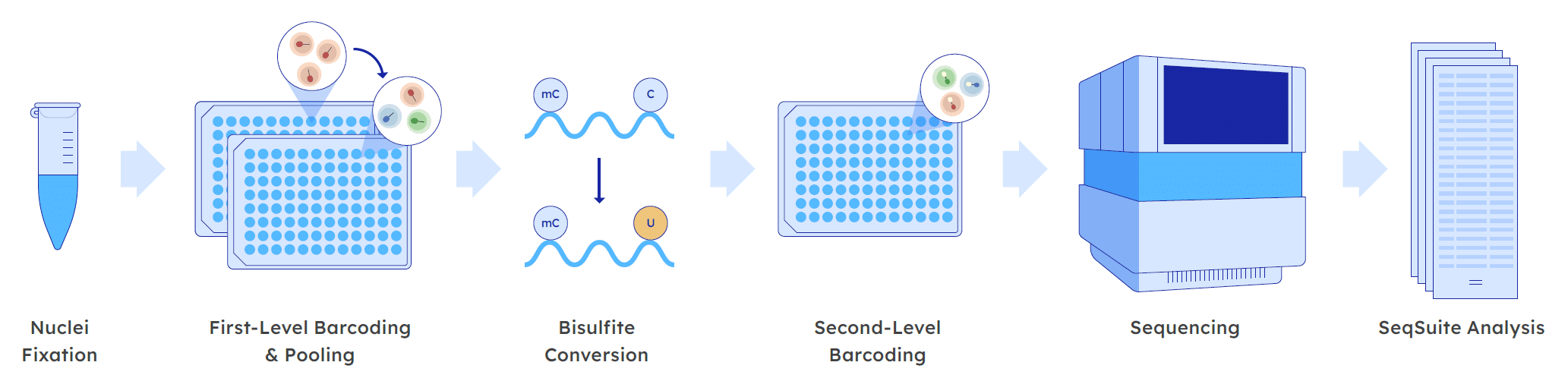
Massively parallelized single cell barcoding, the powerful technology behind Scale Bio’s Single Cell Methylation Sequencing Kit, leverages the cell as the reaction compartment.
-
Fixation and nucleosome depletion: First, nuclei are fixed and nucleosomes depleted. Store for up to 4 weeks, or process right away.
-
First-level indexing: Fixed nuclei are distributed into 96-well plates for in situ tagmentation with a well-specific oligo index. After tagmentation, nuclei are pooled and distributed into a second 96-well plate.
-
Bisulfite conversion: Fixed nuclei undergo bisulfite conversion for detecting methylated cytosines. Unmethylated cytosines are converted to uracils, while methylated cytosines are protected from conversion.
-
Second-level indexing: A second well-specific oligo index is added via PCR, and sequencing adapters are also attached. The combination of the first and second level indices enable a unique cellular barcode for single cell analysis.
-
(Optional) Target enrichment: Using your panel of choice, enrich your libraries using hybridization capture.
-
Sequencing: ScaleBio libraries can be sequenced on several Illumina sequencing platforms, including NextSeq, NovaSeq, and NovaSeqX.
-
Data Analysis: Process raw sequencing data using our SeqSuite analysis pipeline, and easily perform cell calling and generate a single cell methylation matrix. Our tutorials for using secondary analysis software make it easy to go from data to insight.
Featured publications View All
Atlas-scale Single cell DNA Methylation Profiling with sciMETv3
Single cell methods to assess DNA methylation have not yet achieved the same level of cell throughput compared to other modalities.
High-throughput robust single cell DNA methylation profiling with sciMETv2
We demonstrate two versions of sciMETv2 on primary human cortex, a high coverage and rapid version, identifying distinct cell types…
sciMET-cap: high-throughput single cell methylation analysis with a reduced sequencing burden
Leveraging target enrichment with sciMET to capture sufficient information per cell.
Single cell DNA methylation analysis tool Amethyst reveals distinct noncanonical methylation patterns in human glial cells
Here we present Amethyst, a comprehensive R package for atlas-scale single cell methylation sequencing data analysis.
Neurodevelopmental hijacking of oligodendrocyte lineage programs drives glioblastoma infiltration
Performed multi-modal single and spatial profiling of a multi-region GBM patient cohort to characterize infiltrative GBM cells.
Resources View All
New! Join our Methylation Bioinformatics Working Group, meeting monthly. Register
Investigating Lineage-Specific Epigenetic Changes in Human Brain Development
Webinar – Dmitri Velmeshev
Single cell methylome profiling of human glioma
Application Note
Unlock true resolution of Epigenetics with single cell DNA methylation
eBook
Scale Bio Single Cell Methylation Sequencing Kit
Product Sheet
Frequently asked questions
What types of samples have been demonstrated with the Single Cell Methylation Sequencing Kit?
Nuclei from primary tissues in fresh dissociated, snap-frozen, and OCT-embedded formats have been demonstrated in addition to PBMCs and cell lines.
How long can fixed samples be stored for?
Freshly fixed samples can be stored for up to 4 weeks before proceeding to the tagmentation step. Tagmented nuclei can be stored in the freezer for up to 6 months.
What data analysis solutions are provided with the Single Cell Methylation Kit?
A single cell methylation analysis pipeline is provided with the product to take you from raw sequencing outputs to a cell-methylation state matrix.
What sequencers is the kit compatible with?
We have tested compatibility on NextSeq 1000/2000, NovaSeq6000, and NovaSeqX.
Are there any publicly available data sets for methylation?
Yes, you can access our public data sets at the following link: Scale Bio Public Data Sets

What to learn more about our Single Cell Methylation Sequencing Kit?
Reach out to our Team for more information or to request a quote.